(Guest post by Achim Zeileis)
Development of the R package exams for automatic generation of (statistical) exams in R started in 2006 and version 1 was published in JSS by Grün and Zeileis (2009). It was based on standalone Sweave exercises, that can be combined into exams, and then rendered into different kinds of PDF output (exams, solutions, self-study materials, etc.). Now, a major revision of the package has been released that extends the capabilities and adds support for learning management systems. It is still based on the same type of
Sweave files for each exercise but can also render them into output formats like HTML (with various options for displaying mathematical content) and XML specifications for online exams in learning management systems such as Moodle or OLAT. Supplementary files such as graphics or data are
handled automatically. Here, I give a brief overview of the new capabilities. A detailed discussion is in the working paper by Zeileis, Umlauf, and Leisch (2012) that is also contained in the package as a vignette.
Continue reading “Generation of E-Learning Exams in R for Moodle, OLAT, etc.”
Year: 2012
Comparing Shiny with gWidgetsWWW2.rapache
(A guest post by John Verzani)
A few days back the RStudio blog announced Shiny, a new product for easily creating interactive web applications (http://www.rstudio.com/shiny/
I don’t want to worry here about deployment of apps, just the writing side. The shiny package uses websockets to transfer data back and forth from browser to server. Though this may cause issues with wider deployment, the industrious RStudio folks have a hosting program in beta for internet-wide deployment. For local deployment, no problems as far as I know – as long as you avoid older versions of internet explorer.
Now, Shiny seems well suited for applications where the user can parameterize a resulting graphic, so that was the point of comparison. Peter Dalgaard’s tcltk package ships with a classic demo tkdensity.R. I use that for inspiration below. That GUI allows the user a few selections to modify a density plot of a random sample.
Continue reading “Comparing Shiny with gWidgetsWWW2.rapache”
How to load the {rJava} package after the error "JAVA_HOME cannot be determined from the Registry"
In case you tried loading a package that depends on the {rJava} package (by Simon Urbanek), you might came across the following error:
Loading required package: rJava
library(rJava)
Error : .onLoad failed in loadNamespace() for ‘rJava’, details:
call: fun(libname, pkgname)
error: JAVA_HOME cannot be determined from the Registry
The error tells us that there is no entry in the Registry that tells R where Java is located. It is most likely that Java was not installed (or that the registry is corrupt).
This error is often resolved by installing a Java version (i.e. 64-bit Java or 32-bit Java) that fits to the type of R version that you are using (i.e. 64-bit R or 32-bit R). This problem can easily effect Windows 7 users, since they might have installed a version of Java that is different than the version of R they are using.
Note that it is necessary to ‘manually download and install’ the 64 bit version of JAVA. By default, the download page gives a 32 bit version .
You can pick the exact version of Java you wish to install from this link. If you might (for some reason) work on both versions of R, you can install both version of Java (Installing the “Java Runtime Environment” is probably good enough for your needs).
(Source: Uwe Ligges)
Other possible solutions is trying to re-install rJava.
If that doesn’t work, you could also manually set the directory of your Java location by setting it before loading the library:
Sys.setenv(JAVA_HOME='C:\\Program Files\\Java\\jre7') # for 64-bit version
Sys.setenv(JAVA_HOME='C:\\Program Files (x86)\\Java\\jre7') # for 32-bit version
library(rJava)
(Source: “nograpes” from Stackoverflow, which also describes the find.java in the rJava:::.onLoad function)
data.table version 1.8.1 – now allowed numeric columns and big-number (via bit64) in keys!
This is a guest post written by Branson Owen, an enthusiastic R and data.table user.
Wow, a long time desired feature of data.table finally came true in version 1.8.1! data.table now allowed numeric columns and big number (via bit64) in keys! This is quite a big thing to me and I believe to many other R users too. Now I can hardly think any weakiness of data.table. Oh, did I mention it also started to support character column in the keys (rather than coerce to factor)?
For people who are not familiar with but interested in data.table package, data.table is an enhanced data.frame for high-speed indexing, ordered joins, assignment, grouping and list columns in a short and flexible syntax. You can take a look at some task examples here:
News from datatable-help mailing list:
* New functions chmatch() and %chin%, faster versions of match() and %in% for character vectors. They are about 4 times faster than match() on the example in ?chmatch.
* New function set(DT,i,j,value) allows fast assignment to elements of DT.
M = matrix(1,nrow=100000,ncol=100)
DF = as.data.frame(M)
DT = as.data.table(M)
system.time(for (i in 1:1000) DF[i,1L] <- i) # 591.000s
system.time(for (i in 1:1000) DT[i,V1:=i]) # 1.158s
system.time(for (i in 1:1000) M[i,1L] <- i) # 0.016s
system.time(for (i in 1:1000) set(DT,i,1L,i)) # 0.027s
* Numeric columns (type ‘double’) are now allowed in keys and ad hoc by. Other types which use ‘double’ (such as POSIXct and bit64) can now be fully supported.
For advanced and creative users, it also officially supported list columns awhile ago (rather than support it by accident). For example, your column could be a list of vectors, where each of the vector has different length. This can allow very flexible and creative ways to manipulate data.
The code example below use “function column”, i.e. a list of functions
> DT = data.table(ID=1:4,A=rnorm(4),B=rnorm(4),fn=list(min,max))
> str(DT)
Classes ‘data.table’ and 'data.frame': 4 obs. of 4 variables:
$ ID: int 1 2 3 4
$ A : num -0.7135 -2.5217 0.0265 1.0102
$ B : num -0.4116 0.4032 0.1098 0.0669
$ fn:List of 4
..$ :function (..., na.rm = FALSE)
..$ :function (..., na.rm = FALSE)
..$ :function (..., na.rm = FALSE)
..$ :function (..., na.rm = FALSE)
> DT[,fn[[1]](A,B),by=ID]
ID V1
[1,] 1 -0.71352508
[2,] 2 0.40322625
[3,] 3 0.02648949
[4,] 4 1.01022266
[ref] https://r-forge.r-project.org/tracker/index.php?func=detail&aid=1302&group_id=240&atid=978
Speed up your R code using a just-in-time (JIT) compiler
This post is about speeding up your R code using the JIT (just in time) compilation capabilities offered by the new (well, now a year old) {compiler} package. Specifically, dealing with the practical difference between enableJIT and the cmpfun functions.
If you do not want to read much, you can just skip to the example part.
As always, I welcome any comments to this post, and hope to update it when future JIT solutions will come along.
Continue reading “Speed up your R code using a just-in-time (JIT) compiler”
Do more with dates and times in R with lubridate 1.1.0
This is a guest post by Garrett Grolemund (mentored by Hadley Wickham)
Lubridate is an R package that makes it easier to work with dates and times. The newest release of lubridate (v 1.1.0) comes with even more tools and some significant changes over past versions. Below is a concise tour of some of the things lubridate can do for you. At the end of this post, I list some of the differences between lubridate (v 0.2.4) and lubridate (v 1.1.0). If you are an old hand at lubridate, please read this section to avoid surprises!
Lubridate was created by Garrett Grolemund and Hadley Wickham.
Parsing dates and times
Getting R to agree that your data contains the dates and times you think it does can be a bit tricky. Lubridate simplifies that. Identify the order in which the year, month, and day appears in your dates. Now arrange “y”, “m”, and “d” in the same order. This is the name of the function in lubridate that will parse your dates. For example,
library(lubridate)
ymd("20110604"); mdy("06-04-2011"); dmy("04/06/2011")
## "2011-06-04 UTC"
## "2011-06-04 UTC"
## "2011-06-04 UTC"Parsing functions automatically handle a wide variety of formats and separators, which simplifies the parsing process.
If your date includes time information, add h, m, and/or s to the name of the function. ymd_hms() is probably the most common date time format. To read the dates in with a certain time zone, supply the official name of that time zone in the tz argument.
arrive <- ymd_hms("2011-06-04 12:00:00", tz = "Pacific/Auckland")
## "2011-06-04 12:00:00 NZST"
leave <- ymd_hms("2011-08-10 14:00:00", tz = "Pacific/Auckland")
## "2011-08-10 14:00:00 NZST"Setting and Extracting information
Extract information from date times with the functions second(), minute(), hour(), day(), wday(), yday(), week(), month(), year(), and tz(). You can also use each of these to set (i.e, change) the given information. Notice that this will alter the date time. wday() and month() have an optional label argument, which replaces their numeric output with the name of the weekday or month.
second(arrive)
## 0
second(arrive) <- 25
arrive
## "2011-06-04 12:00:25 NZST"
second(arrive) <- 0
wday(arrive)
## 7
wday(arrive, label = TRUE)
## SatTime Zones
There are two very useful things to do with dates and time zones. First, display the same moment in a different time zone. Second, create a new moment by combining a given clock time with a new time zone. These are accomplished by with_tz() and force_tz().
For example, I spent last summer researching in Auckland, New Zealand. I arranged to meet with my advisor, Hadley, over skype at 9:00 in the morning Auckland time. What time was that for Hadley who was back in Houston, TX?
meeting <- ymd_hms("2011-07-01 09:00:00", tz = "Pacific/Auckland")
## "2011-07-01 09:00:00 NZST"
with_tz(meeting, "America/Chicago")
## "2011-06-30 16:00:00 CDT"So the meetings occurred at 4:00 Hadley’s time (and the day before no less). Of course, this was the same actual moment of time as 9:00 in New Zealand. It just appears to be a different day due to the curvature of the Earth.
What if Hadley made a mistake and signed on at 9:00 his time? What time would it then be my time?
mistake <- force_tz(meeting, "America/Chicago")
## "2011-07-01 09:00:00 CDT"
with_tz(mistake, "Pacific/Auckland")
## "2011-07-02 02:00:00 NZST"His call would arrive at 2:00 am my time! Luckily he never did that.
Continue reading "Do more with dates and times in R with lubridate 1.1.0"
Printing nested tables in R – bridging between the {reshape} and {tables} packages
This post shows how to print a prettier nested pivot table, created using the {reshape} package (similar to what you would get with Microsoft Excel), so you could print it either in the R terminal or as a LaTeX table. This task is done by bridging between the cast_df object produced by the {reshape} package, […]
This post shows how to print a prettier nested pivot table, created using the {reshape} package (similar to what you would get with Microsoft Excel), so you could print it either in the R terminal or as a LaTeX table. This task is done by bridging between the cast_df object produced by the {reshape} package, and the tabular function introduced by the new {tables} package.
Here is an example of the type of output we wish to produce in the R terminal:
1 2 3 4 5 6 7 | ozone solar.r wind temp month mean sd mean sd mean sd mean sd 5 23.62 22.22 181.3 115.08 11.623 3.531 65.55 6.855 6 29.44 18.21 190.2 92.88 10.267 3.769 79.10 6.599 7 59.12 31.64 216.5 80.57 8.942 3.036 83.90 4.316 8 59.96 39.68 171.9 76.83 8.794 3.226 83.97 6.585 9 31.45 24.14 167.4 79.12 10.180 3.461 76.90 8.356 |
Or in a latex document:
Motivation: creating pretty nested tables
In a recent post we learned how to use the {reshape} package (by Hadley Wickham) in order to aggregate and reshape data (in R) using the melt and cast functions.
The cast function is wonderful but it has one problem – the format of the output. As opposed to a pivot table in (for example) MS excel, the output of a nested table created by cast is very “flat”. That is, there is only one row for the header, and only one column for the row names. So for both the R terminal, or an Sweave document, when we deal with a more complex reshaping/aggregating, the result is not something you would be proud to send to a journal.
The opportunity: the {tables} package
The good news is that Duncan Murdoch have recently released a new package to CRAN called {tables}. The {tables} package can compute and display complex tables of summary statistics and turn them into nice looking tables in Sweave (LaTeX) documents. For using the full power of this package, you are invited to read through its detailed (and well written) 23 pages Vignette. However, some of us might have preferred to keep using the syntax of the {reshape} package, while also benefiting from the great formatting that is offered by the new {tables} package. For this purpose, I devised a function that bridges between cast_df (from {reshape}) and the tabular function (from {tables}).
The bridge: between the {tables} and the {reshape} packages
The code for the function is available on my github (link: tabular.cast_df.r on github) and it seems to works fine as far as I can see (though I wouldn’t run it on larger data files since it relies on melting a cast_df object.)
Here is an example for how to load and use the function:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 | ###################### # Loading the functions ###################### # Making sure we can source code from github source("https://www.r-statistics.com/wp-content/uploads/2012/01/source_https.r.txt") # Reading in the function for using tabular on a cast_df object: source_https("https://raw.github.com/talgalili/R-code-snippets/master/tabular.cast_df.r") ###################### # example: ###################### ############ # Loading and preparing some data require(reshape) names(airquality) <- tolower(names(airquality)) airquality2 <- airquality airquality2$temp2 <- ifelse(airquality2$temp > median(airquality2$temp), "hot", "cold") aqm <- melt(airquality2, id=c("month", "day","temp2"), na.rm=TRUE) colnames(aqm)[4] <- "variable2" # because otherwise the function is having problem when relying on the melt function of the cast object head(aqm,3) # month day temp2 variable2 value #1 5 1 cold ozone 41 #2 5 2 cold ozone 36 #3 5 3 cold ozone 12 ############ # Running the example: tabular.cast_df(cast(aqm, month ~ variable2, c(mean,sd))) tabular(cast(aqm, month ~ variable2, c(mean,sd))) # notice how we turned tabular to be an S3 method that can deal with a cast_df object Hmisc::latex(tabular(cast(aqm, month ~ variable2, c(mean,sd)))) # this is what we would have used for an Sweave document |
And here are the results in the terminal:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | > > tabular.cast_df(cast(aqm, month ~ variable2, c(mean,sd))) ozone solar.r wind temp month mean sd mean sd mean sd mean sd 5 23.62 22.22 181.3 115.08 11.623 3.531 65.55 6.855 6 29.44 18.21 190.2 92.88 10.267 3.769 79.10 6.599 7 59.12 31.64 216.5 80.57 8.942 3.036 83.90 4.316 8 59.96 39.68 171.9 76.83 8.794 3.226 83.97 6.585 9 31.45 24.14 167.4 79.12 10.180 3.461 76.90 8.356 > tabular(cast(aqm, month ~ variable2, c(mean,sd))) # notice how we turned tabular to be an S3 method that can deal with a cast_df object ozone solar.r wind temp month mean sd mean sd mean sd mean sd 5 23.62 22.22 181.3 115.08 11.623 3.531 65.55 6.855 6 29.44 18.21 190.2 92.88 10.267 3.769 79.10 6.599 7 59.12 31.64 216.5 80.57 8.942 3.036 83.90 4.316 8 59.96 39.68 171.9 76.83 8.794 3.226 83.97 6.585 9 31.45 24.14 167.4 79.12 10.180 3.461 76.90 8.356 |
And in an Sweave document:
Here is an example for the Rnw file that produces the above table:
cast_df to tabular.Rnw
I will finish with saying that the tabular function offers more flexibility then the one offered by the function I provided. If you find any bugs or have suggestions of improvement, you are invited to leave a comment here or inside the code on github.
(Link-tip goes to Tony Breyal for putting together a solution for sourcing r code from github.)
Interactive Graphics with the iplots Package (from “R in Action”)
The followings introductory post is intended for new users of R. It deals with interactive visualization using R through the iplots package.
This is a guest article by Dr. Robert I. Kabacoff, the founder of (one of) the first online R tutorials websites: Quick-R. Kabacoff has recently published the book ”R in Action“, providing a detailed walk-through for the R language based on various examples for illustrating R’s features (data manipulation, statistical methods, graphics, and so on…). In previous guest posts by Kabacoff we introduced data.frame objects in R and dealt with the Aggregation and Restructuring of data (using base R functions and the reshape package).
For readers of this blog, there is a 38% discount off the “R in Action” book (as well as all other eBooks, pBooks and MEAPs at Manning publishing house), simply by using the code rblogg38 when reaching checkout.
Let us now talk about Interactive Graphics with the iplots Package:

Interactive Graphics with the iplots Package
The base installation of R provides limited interactivity with graphs. You can modify graphs by issuing additional program statements, but there’s little that you can do to modify them or gather new information from them using the mouse. However, there are contributed packages that greatly enhance your ability to interact with the graphs you create—playwith, latticist, iplots, and rggobi. In this article, we’ll focus on functions provided by the iplots package. Be sure to install it before first use.
While playwith and latticist allow you to interact with a single graph, the iplots package takes interaction in a different direction. This package provides interactive mosaic plots, bar plots, box plots, parallel plots, scatter plots, and histograms that can be linked together and color brushed. This means that you can select and identify observations using the mouse, and highlighting observations in one graph will automatically highlight the same observations in all other open graphs. You can also use the mouse to obtain information about graphic objects such as points, bars, lines, and box plots.
The iplots package is implemented through Java and the primary functions are listed in table 1.
Table 1 iplot functions
Function | Description |
| ibar() | Interactive bar chart |
| ibox() | Interactive box plot |
| ihist() | Interactive histogram |
| imap() | Interactive map |
| imosaic() | Interactive mosaic plot |
| ipcp() | Interactive parallel coordinates plot |
| iplot() | Interactive scatter plot |
To understand how iplots works, execute the code provided in listing 1.
Listing 1 iplots demonstration
1 2 3 4 5 6 7 8 9 10 11 12 | library(iplots) attach(mtcars) cylinders <- factor(cyl) gears <- factor(gear) transmission <- factor(am) ihist(mpg) ibar(gears) iplot(mpg, wt) ibox(mtcars[c("mpg", "wt", "qsec", "disp", "hp")]) ipcp(mtcars[c("mpg", "wt", "qsec", "disp", "hp")]) imosaic(transmission, cylinders) detach(mtcars) |
Six windows containing graphs will open. Rearrange them on the desktop so that each is visible (each can be resized if necessary). A portion of the display is provided in figure 1.
Figure 1 An iplots demonstration created by listing 1. Only four of the six windows are displayed to save room. In these graphs, the user has clicked on the three-gear bar in the bar chart window.
Now try the following:
- Click on the three-gear bar in the Barchart (gears) window. The bar will turn red. In addition, all cars with three-gear engines will be highlighted in the other graph windows.
- Mouse down and drag to select a rectangular region of points in the Scatter plot (wt vs mpg) window. These points will be highlighted and the corresponding observations in every other graph window will also turn red.
- Hold down the Ctrl key and move the mouse pointer over a point, bar, box plot, or line in one of the graphs. Details about that object will appear in a pop-up window.
- Right-click on any object and note the options that are offered in the context menu. For example, you can right-click on the Boxplot (mpg) window and change the graph to a parallel coordinates plot (PCP).
- You can drag to select more than one object (point, bar, and so on) or use Shift-click to select noncontiguous objects. Try selecting both the three- and five-gear bars in the Barchart (gears) window.
The functions in the iplots package allow you to explore the variable distributions and relationships among variables in subgroups of observations that you select interactively. This can provide insights that would be difficult and time-consuming to obtain in other ways. For more information on the iplots package, visit the project website at http://rosuda.org/iplots/.
Summary
In this article, we explored one of the several packages for dynamically interacting with graphs, iplots. This package allows you to interact directly with data in graphs, leading to a greater intimacy with your data and expanded opportunities for developing insights.
This article first appeared as chapter 16.4.4 from the “R in action“ book, and is published with permission from Manning publishing house. Other books in this serious which you might be interested in are (see the beginning of this post for a discount code):
- Machine Learning in Action by Peter Harrington
- Gnuplot in Action (Understanding Data with Graphs) by Philipp K. Janert
Merging two data.frame objects while preserving the rows’ order
Merging two data.frame objects in R is very easily done by using the merge function. While being very powerful, the merge function does not (as of yet) offer to return a merged data.frame that preserved the original order of, one of the two merged, data.frame objects. In this post I describe this problem, and offer […]
Update (2017-02-03) the dplyr package offers a great solution for this issue, see the document Two-table verbs for more details.
Merging two data.frame objects in R is very easily done by using the merge function. While being very powerful, the merge function does not (as of yet) offer to return a merged data.frame that preserved the original order of, one of the two merged, data.frame objects.
In this post I describe this problem, and offer some easy to use code to solve it.
Let us start with a simple example:
x <- data.frame( ref = c( 'Ref1', 'Ref2' ) , label = c( 'Label01', 'Label02' ) ) y <- data.frame( id = c( 'A1', 'C2', 'B3', 'D4' ) , ref = c( 'Ref1', 'Ref2' , 'Ref3','Ref1' ) , val = c( 1.11, 2.22, 3.33, 4.44 ) ) ####################### # having a look at the two data.frame objects: > x ref label 1 Ref1 Label01 2 Ref2 Label02 > y id ref val 1 A1 Ref1 1.11 2 C2 Ref2 2.22 3 B3 Ref3 3.33 4 D4 Ref1 4.44 |
If we will now merge the two objects, we will find that the order of the rows is different then the original order of the “y” object. This is true whether we use “sort =T” or “sort=F”. You can notice that the original order was an ascending order of the “val” variable:
> merge( x, y, by='ref', all.y = T, sort= T) ref label id val 1 Ref1 Label01 A1 1.11 2 Ref1 Label01 D4 4.44 3 Ref2 Label02 C2 2.22 4 Ref3 <NA> B3 3.33 > merge( x, y, by='ref', all.y = T, sort=F ) ref label id val 1 Ref1 Label01 A1 1.11 2 Ref1 Label01 D4 4.44 3 Ref2 Label02 C2 2.22 4 Ref3 <NA> B3 3.33 |
This is explained in the help page of ?merge:
The rows are by default lexicographically sorted on the common columns, but for ‘sort = FALSE’ are in an unspecified order.
Or put differently: sort=FALSE doesn’t preserve the order of any of the two entered data.frame objects (x or y); instead it gives us an
unspecified (potentially random) order.
However, it can so happen that we want to make sure the order of the resulting merged data.frame objects ARE ordered according to the order of one of the two original objects. In order to make sure of that, we could add an extra “id” (row index number) sequence on the dataframe we wish to sort on. Then, we can merge the two data.frame objects, sort by the sequence, and delete the sequence. (this was previously mentioned on the R-help mailing list by Bart Joosen).
Following is a function that implements this logic, followed by an example for its use:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 | ############## function: merge.with.order <- function(x,y, ..., sort = T, keep_order) { # this function works just like merge, only that it adds the option to return the merged data.frame ordered by x (1) or by y (2) add.id.column.to.data <- function(DATA) { data.frame(DATA, id... = seq_len(nrow(DATA))) } # add.id.column.to.data(data.frame(x = rnorm(5), x2 = rnorm(5))) order.by.id...and.remove.it <- function(DATA) { # gets in a data.frame with the "id..." column. Orders by it and returns it if(!any(colnames(DATA)=="id...")) stop("The function order.by.id...and.remove.it only works with data.frame objects which includes the 'id...' order column") ss_r <- order(DATA$id...) ss_c <- colnames(DATA) != "id..." DATA[ss_r, ss_c] } # tmp <- function(x) x==1; 1 # why we must check what to do if it is missing or not... # tmp() if(!missing(keep_order)) { if(keep_order == 1) return(order.by.id...and.remove.it(merge(x=add.id.column.to.data(x),y=y,..., sort = FALSE))) if(keep_order == 2) return(order.by.id...and.remove.it(merge(x=x,y=add.id.column.to.data(y),..., sort = FALSE))) # if you didn't get "return" by now - issue a warning. warning("The function merge.with.order only accepts NULL/1/2 values for the keep_order variable") } else {return(merge(x=x,y=y,..., sort = sort))} } ######### example: > merge( x.labels, x.vals, by='ref', all.y = T, sort=F ) ref label id val 1 Ref1 Label01 A1 1.11 2 Ref1 Label01 D4 4.44 3 Ref2 Label02 C2 2.22 4 Ref3 <NA> B3 3.33 > merge.with.order( x.labels, x.vals, by='ref', all.y = T, sort=F ,keep_order = 1) ref label id val 1 Ref1 Label01 A1 1.11 2 Ref1 Label01 D4 4.44 3 Ref2 Label02 C2 2.22 4 Ref3 <NA> B3 3.33 > merge.with.order( x.labels, x.vals, by='ref', all.y = T, sort=F ,keep_order = 2) # yay - works as we wanted it to... ref label id val 1 Ref1 Label01 A1 1.11 3 Ref2 Label02 C2 2.22 4 Ref3 <NA> B3 3.33 2 Ref1 Label01 D4 4.44 |
Here is a description for how to use the keep_order parameter:
keep_order can accept the numbers 1 or 2, in which case it will make sure the resulting merged data.frame will be ordered according to the original order of rows of the data.frame entered to x (if keep_order=1) or to y (if keep_order=2). If keep_order is missing, merge will continue working as usual. If keep_order gets some input other then 1 or 2, it will issue a warning that it doesn’t accept these values, but will continue working as merge normally would. Notice that the parameter “sort” is practically overridden when using keep_order (with the value 1 or 2).
The same code can be used to modify the original merge.data.frame function in base R, so to allow the use of the keep_order, here is a link to the patched merge.data.frame function (on github). If you can think of any ways to improve the function (or happen to notice a bug) please let me know either on github or in the comments. (also saying that you found the function to be useful will be fun to know about ![]() )
)
Update: Thanks to KY’s comment, I noticed the ?join function in the {plyr} library. This function is similar to merge (with less features, yet faster), and also automatically keeps the order of the x (first) data.frame used for merging, as explained in the ?join help page:
Unlike merge, (join) preserves the order of x no matter what join type is used. If needed, rows from y will be added to the bottom. Join is often faster than merge, although it is somewhat less featureful – it currently offers no way to rename output or merge on different variables in the x and y data frames.
Aggregation and Restructuring data (from “R in Action”)
The followings introductory post is intended for new users of R. It deals with the restructuring of data: what it is and how to perform it using base R functions and the {reshape} package. This is a guest article by Dr. Robert I. Kabacoff, the founder of (one of) the first online R tutorials websites: Quick-R. Kabacoff […]
The followings introductory post is intended for new users of R. It deals with the restructuring of data: what it is and how to perform it using base R functions and the {reshape} package.
This is a guest article by Dr. Robert I. Kabacoff, the founder of (one of) the first online R tutorials websites: Quick-R. Kabacoff has recently published the book ”R in Action“, providing a detailed walk-through for the R language based on various examples for illustrating R’s features (data manipulation, statistical methods, graphics, and so on…). The previous guest post by Kabacoff introduced data.frame objects in R.

Let us now talk about the Aggregation and Restructuring of data in R:

Aggregation and Restructuring
R provides a number of powerful methods for aggregating and reshaping data. When you aggregate data, you replace groups of observations with summary statistics based on those observations. When you reshape data, you alter the structure (rows and columns) determining how the data is organized. This article describes a variety of methods for accomplishing these tasks.
We’ll use the mtcars data frame that’s included with the base installation of R. This dataset, extracted from Motor Trend magazine (1974), describes the design and performance characteristics (number of cylinders, displacement, horsepower, mpg, and so on) for 34 automobiles. To learn more about the dataset, see help(mtcars).
Transpose
The transpose (reversing rows and columns) is perhaps the simplest method of reshaping a dataset. Use the t() function to transpose a matrix or a data frame. In the latter case, row names become variable (column) names. An example is presented in the next listing.
Listing 1 Transposing a dataset
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | > cars <- mtcars[1:5,1:4] > cars mpg cyl disp hp Mazda RX4 21.0 6 160 110 Mazda RX4 Wag 21.0 6 160 110 Datsun 710 22.8 4 108 93 Hornet 4 Drive 21.4 6 258 110 Hornet Sportabout 18.7 8 360 175 > t(cars) Mazda RX4 Mazda RX4 Wag Datsun 710 Hornet 4 Drive Hornet Sportabout mpg 21 21 22.8 21.4 18.7 cyl 6 6 4.0 6.0 8.0 disp 160 160 108.0 258.0 360.0 hp 110 110 93.0 110.0 175.0 |
Listing 1 uses a subset of the mtcars dataset in order to conserve space on the page. You’ll see a more flexible way of transposing data when we look at the reshape package later in this article.
Aggregating data
It’s relatively easy to collapse data in R using one or more by variables and a defined function. The format is
1 | aggregate(x, by, FUN) |
where x is the data object to be collapsed, by is a list of variables that will be crossed to form the new observations, and FUN is the scalar function used to calculate summary statistics that will make up the new observation values.
As an example, we’ll aggregate the mtcars data by number of cylinders and gears, returning means on each of the numeric variables (see the next listing).
Listing 2 Aggregating data
1 2 3 4 5 6 7 8 9 10 11 12 13 | > options(digits=3) > attach(mtcars) > aggdata <-aggregate(mtcars, by=list(cyl,gear), FUN=mean, na.rm=TRUE) > aggdata Group.1 Group.2 mpg cyl disp hp drat wt qsec vs am gear carb 1 4 3 21.5 4 120 97 3.70 2.46 20.0 1.0 0.00 3 1.00 2 6 3 19.8 6 242 108 2.92 3.34 19.8 1.0 0.00 3 1.00 3 8 3 15.1 8 358 194 3.12 4.10 17.1 0.0 0.00 3 3.08 4 4 4 26.9 4 103 76 4.11 2.38 19.6 1.0 0.75 4 1.50 5 6 4 19.8 6 164 116 3.91 3.09 17.7 0.5 0.50 4 4.00 6 4 5 28.2 4 108 102 4.10 1.83 16.8 0.5 1.00 5 2.00 7 6 5 19.7 6 145 175 3.62 2.77 15.5 0.0 1.00 5 6.00 8 8 5 15.4 8 326 300 3.88 3.37 14.6 0.0 1.00 5 6.00 |
In these results, Group.1 represents the number of cylinders (4, 6, or ![]() and Group.2 represents the number of gears (3, 4, or 5). For example, cars with 4 cylinders and 3 gears have a mean of 21.5 miles per gallon (mpg).
and Group.2 represents the number of gears (3, 4, or 5). For example, cars with 4 cylinders and 3 gears have a mean of 21.5 miles per gallon (mpg).
When you’re using the aggregate() function , the by variables must be in a list (even if there’s only one). You can declare a custom name for the groups from within the list, for instance, using by=list(Group.cyl=cyl, Group.gears=gear).
The function specified can be any built-in or user-provided function. This gives the aggregate command a great deal of power. But when it comes to power, nothing beats the reshape package.
The reshape package
The reshape package is a tremendously versatile approach to both restructuring and aggregating datasets. Because of this versatility, it can be a bit challenging to learn.
We’ll go through the process slowly and use a small dataset so that it’s clear what’s happening. Because reshape isn’t included in the standard installation of R, you’ll need to install it one time, using install.packages(“reshape”).
Basically, you’ll “melt” data so that each row is a unique ID-variable combination. Then you’ll “cast” the melted data into any shape you desire. During the cast, you can aggregate the data with any function you wish. The dataset you’ll be working with is shown in table 1.
Table 1 The original dataset (mydata)
ID | Time | X1 | X2 |
| 1 | 1 | 5 | 6 |
| 1 | 2 | 3 | 5 |
| 2 | 1 | 6 | 1 |
| 2 | 2 | 2 | 4 |
In this dataset, the measurements are the values in the last two columns (5, 6, 3, 5, 6, 1, 2, and 4). Each measurement is uniquely identified by a combination of ID variables (in this case ID, Time, and whether the measurement is on X1 or X2). For example, the measured value 5 in the first row is uniquely identified by knowing that it’s from observation (ID) 1, at Time 1, and on variable X1.
Melting
When you melt a dataset, you restructure it into a format where each measured variable is in its own row, along with the ID variables needed to uniquely identify it. If you melt the data from table 1, using the following code
1 2 | library(reshape) md <- melt(mydata, id=(c("id", "time"))) |
You end up with the structure shown in table 2.
Table 2 The melted dataset
ID | Time | Variable | Value |
| 1 | 1 | X1 | 5 |
| 1 | 2 | X1 | 3 |
| 2 | 1 | X1 | 6 |
| 2 | 2 | X1 | 2 |
| 1 | 1 | X2 | 6 |
| 1 | 2 | X2 | 5 |
| 2 | 1 | X2 | 1 |
| 2 | 2 | X2 | 4 |
Note that you must specify the variables needed to uniquely identify each measurement (ID and Time) and that the variable indicating the measurement variable names (X1 or X2) is created for you automatically.
Now that you have your data in a melted form, you can recast it into any shape, using the cast() function.
Casting
The cast() function starts with melted data and reshapes it using a formula that you provide and an (optional) function used to aggregate the data. The format is
1 | newdata <- cast(md, formula, FUN) |
Where md is the melted data, formula describes the desired end result, and FUN is the (optional) aggregating function. The formula takes the form
1 | rowvar1 + rowvar2 + … ~ colvar1 + colvar2 + … |
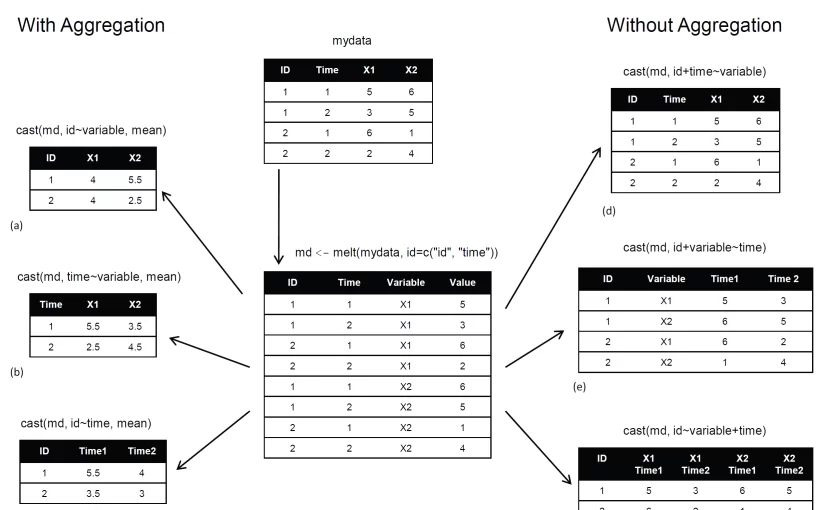
In this formula, rowvar1 + rowvar2 + … define the set of crossed variables that define the rows, and colvar1 + colvar2 + … define the set of crossed variables that define the columns. See the examples in figure 1. (click to enlarge the image)
Figure 1 Reshaping data with the melt() and cast() functions
Because the formulas on the right side (d, e, and f) don’t include a function, the data is reshaped. In contrast, the examples on the left side (a, b, and c) specify the mean as an aggregating function. Thus the data are not only reshaped but aggregated as well. For example, (a) gives the means on X1 and X2 averaged over time for each observation. Example (b) gives the mean scores of X1 and X2 at Time 1 and Time 2, averaged over observations. In (c) you have the mean score for each observation at Time 1 and Time 2, averaged over X1 and X2.
As you can see, the flexibility provided by the melt() and cast() functions is amazing. There are many times when you’ll have to reshape or aggregate your data prior to analysis. For example, you’ll typically need to place your data in what’s called long format resembling table 2 when analyzing repeated measures data (data where multiple measures are recorded for each observation).
Summary
Chapter 5 of R in Action reviews many of the dozens of mathematical, statistical, and probability functions that are useful for manipulating data. In this article, we have briefly explored several ways of aggregating and restructuring data.
This article first appeared as chapter 5.6 from the “R in action“ book, and is published with permission from Manning publishing house. Other books in this serious which you might be interested in are (see the beginning of this post for a discount code):
- Machine Learning in Action by Peter Harrington
- Gnuplot in Action (Understanding Data with Graphs) by Philipp K. Janert