About Clustergrams
In 2002, Matthias Schonlau published in “The Stata Journal” an article named “The Clustergram: A graph for visualizing hierarchical and . As explained in the abstract:
In hierarchical cluster analysis dendrogram graphs are used to visualize how clusters are formed. I propose an alternative graph named “clustergram” to examine how cluster members are assigned to clusters as the number of clusters increases.
This graph is useful in exploratory analysis for non-hierarchical clustering algorithms like k-means and for hierarchical cluster algorithms when the number of observations is large enough to make dendrograms impractical.
A similar article was later written and was (maybe) published in “computational statistics”.
Both articles gives some nice background to known methods like k-means and methods for hierarchical clustering, and then goes on to present examples of using these methods (with the Clustergarm) to analyse some datasets.
Personally, I understand the clustergram to be a type of parallel coordinates plot where each observation is given a vector. The vector contains the observation’s location according to how many clusters the dataset was split into. The scale of the vector is the scale of the first principal component of the data.
Clustergram in R (a basic function)
After finding out about this method of visualization, I was hunted by the curiosity to play with it a bit. Therefore, and since I didn’t find any implementation of the graph in R, I went about writing the code to implement it.
The code only works for kmeans, but it shows how such a plot can be produced, and could be later modified so to offer methods that will connect with different clustering algorithms.
How does the function work: The function I present here gets a data.frame/matrix with a row for each observation, and the variable dimensions present in the columns.
The function assumes the data is scaled.
The function then goes about calculating the cluster centers for our data, for varying number of clusters.
For each cluster iteration, the cluster centers are multiplied by the first loading of the principal components of the original data. Thus offering a weighted mean of the each cluster center dimensions that might give a decent representation of that cluster (this method has the known limitations of using the first component of a PCA for dimensionality reduction, but I won’t go into that in this post).
Finally all of our data points are ordered according to their respective cluster first component, and plotted against the number of clusters (thus creating the clustergram).
My thank goes to Hadley Wickham for offering some good tips on how to prepare the graph.
Here is the code (example follows)
The R function can be downloaded from here
Corrections and remarks can be added in the comments bellow, or on the github code page.
Example on the iris dataset
The iris data set is a favorite example of many R bloggers when writing about R accessors , Data Exporting, Data importing, and for different visualization techniques.
So it seemed only natural to experiment on it here.
source("https://www.r-statistics.com/wp-content/uploads/2012/01/source_https.r.txt") # Making sure we can source code from github
source_https("https://raw.github.com/talgalili/R-code-snippets/master/clustergram.r")
data(iris)
set.seed(250)
par(cex.lab = 1.5, cex.main = 1.2)
Data <- scale(iris[,-5]) # notice I am scaling the vectors)
clustergram(Data, k.range = 2:8, line.width = 0.004) # notice how I am using line.width. Play with it on your problem, according to the scale of Y.
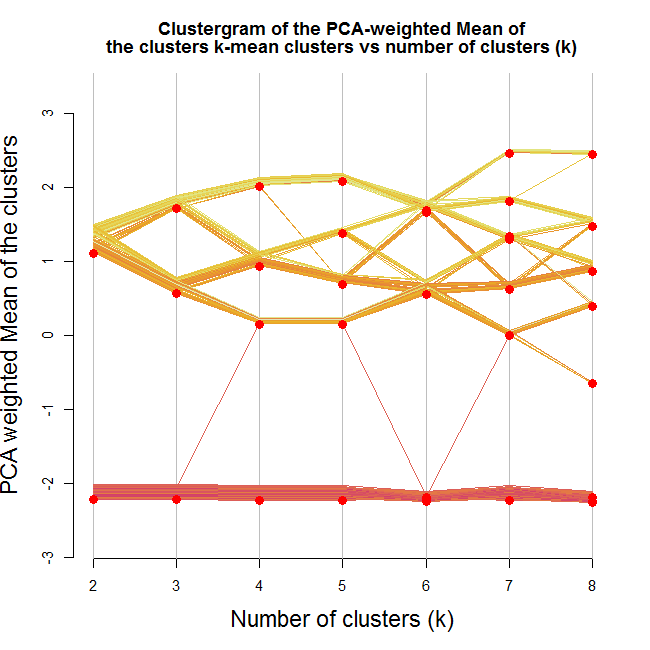
Here is the output:
Looking at the image we can notice a few interesting things. We notice that one of the clusters formed (the lower one) stays as is no matter how many clusters we are allowing (except for one observation that goes way and then beck).
We can also see that the second split is a solid one (in the sense that it splits the first cluster into two clusters which are not "close" to each other, and that about half the observations goes to each of the new clusters).
And then notice how moving to 5 clusters makes almost no difference.
Lastly, notice how when going for 8 clusters, we are practically left with 4 clusters (remember - this is according the mean of cluster centers by the loading of the first component of the PCA on the data)
If I where to take something from this graph, I would say I have a strong tendency to use 3-4 clusters on this data.
But wait, did our clustering algorithm do a stable job?
Let's try running the algorithm 6 more times (each run will have a different starting point for the clusters)
source("https://www.r-statistics.com/wp-content/uploads/2012/01/source_https.r.txt") # Making sure we can source code from github
source_https("https://raw.github.com/talgalili/R-code-snippets/master/clustergram.r")
set.seed(500)
Data <- scale(iris[,-5]) # notice I am scaling the vectors)
par(cex.lab = 1.2, cex.main = .7)
par(mfrow = c(3,2))
for(i in 1:6) clustergram(Data, k.range = 2:8 , line.width = .004, add.center.points = T)
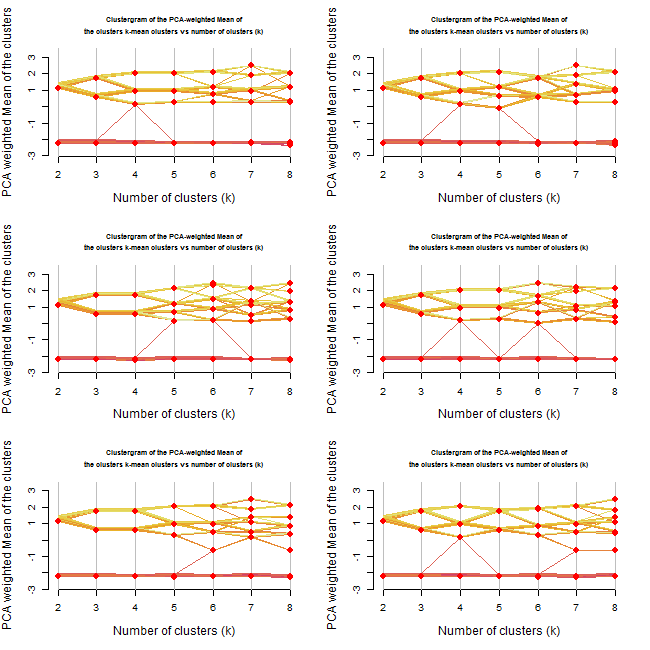
Resulting with: (press the image to enlarge it)
Repeating the analysis offers even more insights.
First, it would appear that until 3 clusters, the algorithm gives rather stable results.
From 4 onwards we get various outcomes at each iteration.
At some of the cases, we got 3 clusters when we asked for 4 or even 5 clusters.
Reviewing the new plots, I would prefer to go with the 3 clusters option. Noting how the two "upper" clusters might have similar properties while the lower cluster is quite distinct from the other two.
By the way, the Iris data set is composed of three types of flowers. I imagine the kmeans had done a decent job in distinguishing the three.
Limitation of the method (and a possible way to overcome it?!)
It is worth noting that the current way the algorithm is built has a fundamental limitation: The plot is good for detecting a situation where there are several clusters but each of them is clearly "bigger" then the one before it (on the first principal component of the data).
For example, let's create a dataset with 3 clusters, each one is taken from a normal distribution with a higher mean:
source("https://www.r-statistics.com/wp-content/uploads/2012/01/source_https.r.txt") # Making sure we can source code from github
source_https("https://raw.github.com/talgalili/R-code-snippets/master/clustergram.r")
set.seed(250)
Data <- rbind(
cbind(rnorm(100,0, sd = 0.3),rnorm(100,0, sd = 0.3),rnorm(100,0, sd = 0.3)),
cbind(rnorm(100,1, sd = 0.3),rnorm(100,1, sd = 0.3),rnorm(100,1, sd = 0.3)),
cbind(rnorm(100,2, sd = 0.3),rnorm(100,2, sd = 0.3),rnorm(100,2, sd = 0.3))
)
clustergram(Data, k.range = 2:5 , line.width = .004, add.center.points = T)
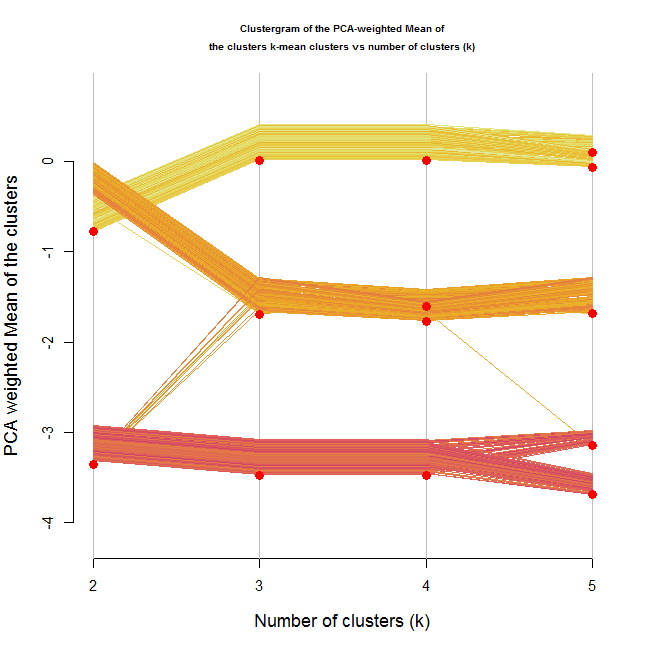
The resulting plot for this is the following:
The image shows a clear distinction between three ranks of clusters. There is no doubt (for me) from looking at this image, that three clusters would be the correct number of clusters.
But what if the clusters where different but didn't have an ordering to them?
For example, look at the following 4 dimensional data:
source("https://www.r-statistics.com/wp-content/uploads/2012/01/source_https.r.txt") # Making sure we can source code from github
source_https("https://raw.github.com/talgalili/R-code-snippets/master/clustergram.r")
set.seed(250)
Data <- rbind(
cbind(rnorm(100,1, sd = 0.3),rnorm(100,0, sd = 0.3),rnorm(100,0, sd = 0.3),rnorm(100,0, sd = 0.3)),
cbind(rnorm(100,0, sd = 0.3),rnorm(100,1, sd = 0.3),rnorm(100,0, sd = 0.3),rnorm(100,0, sd = 0.3)),
cbind(rnorm(100,0, sd = 0.3),rnorm(100,1, sd = 0.3),rnorm(100,1, sd = 0.3),rnorm(100,0, sd = 0.3)),
cbind(rnorm(100,0, sd = 0.3),rnorm(100,0, sd = 0.3),rnorm(100,0, sd = 0.3),rnorm(100,1, sd = 0.3))
)
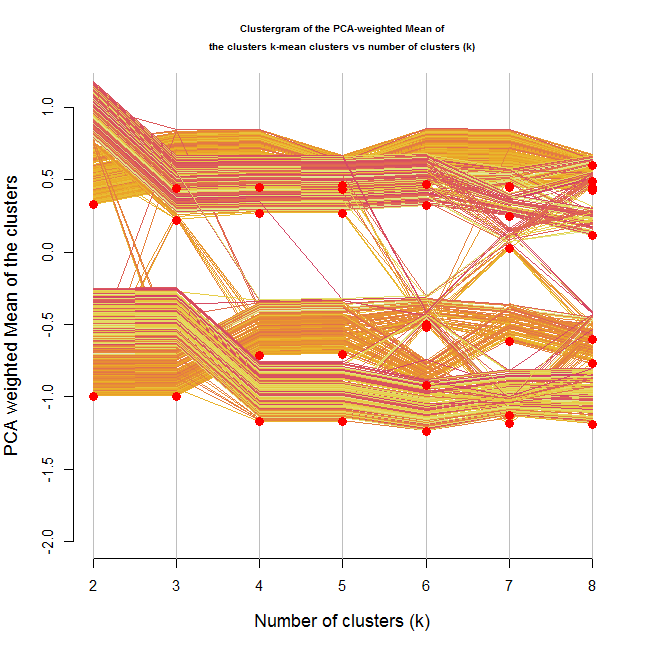
clustergram(Data, k.range = 2:8 , line.width = .004, add.center.points = T)
In this situation, it is not clear from the location of the clusters on the Y axis that we are dealing with 4 clusters.
But what is interesting, is that through the growing number of clusters, we can notice that there are 4 "strands" of data points moving more or less together (until we reached 4 clusters, at which point the clusters started breaking up).
Another hope for handling this might be using the color of the lines in some way, but I haven't yet figured out how.
Clustergram with ggplot2
Hadley Wickham has kindly played with recreating the clustergram using the ggplot2 engine. You can see the result here:
http://gist.github.com/439761
And this is what he wrote about it in the comments:
I’ve broken it down into three components:
* run the clustering algorithm and get predictions (many_kmeans and all_hclust)
* produce the data for the clustergram (clustergram)
* plot it (plot.clustergram)
I don’t think I have the logic behind the y-position adjustment quite right though.
Conclusions (some rules of thumb and questions for the future)
In a first look, it would appear that the clustergram can be of use. I can imagine using this graph to quickly run various clustering algorithms and then compare them to each other and review their stability (In the way I just demonstrated in the example above).
The three rules of thumb I have noticed by now are:
- Look at the location of the cluster points on the Y axis. See when they remain stable, when they start flying around, and what happens to them in higher number of clusters (do they re-group together)
- Observe the strands of the datapoints. Even if the clusters centers are not ordered, the lines for each item might (needs more research and thinking) tend to move together - hinting at the real number of clusters
- Run the plot multiple times to observe the stability of the cluster formation (and location)
Yet there is more work to be done and questions to seek answers to:
- The code needs to be extended to offer methods to various clustering algorithms.
- How can the colors of the lines be used better?
- How can this be done using other graphical engines (ggplot2/lattice?) - (Update: look at Hadley's reply in the comments)
- What to do in case the first principal component doesn't capture enough of the data? (maybe plot this graph to all the relevant components. but then - how do you make conclusions of it?)
- What other uses/conclusions can be made based on this graph?
I am looking forward to reading your input/ideas in the comments (or in reply posts).
I like the idea but I am not sure how to apply it generally. How about the data set from the k-means discussion in MASS:
swiss.x <- scale(as.matrix(swiss[, -1])) # Following your use of scale()
# Not sure:
set.seed(250); clustergram(swiss.x, k.range = 2:10)
# This looks (much) cleaner, but the approach is very seed dependent?:
set.seed(1); clustergram(swiss.x, k.range = 2:10)
Hi Allan,
Here is the code I used for the MASS swiss Dataset:
Two things to consider regarding the MASS swiss example:
1) Notice that because the Y scales are different then in the example I gave in the post, we needed a different parameter for line.width
2) I re-run your example with 6 plots. Looking at it does show that the kmeans is doing a stable (good?!) job with the first two clusters. And that it goes more or less with 3 steady clusters (except in the second plot).
After writing this, I looked at the MASS book (page 317) where he uses the swiss dataset. I can see that it is indeed not a trivial dataset to interpret, and that in the book he went with 3 clusters (so that gives me some good feeling regarding the way I interpreted the clustergram)
I hope what I wrote helped 🙂
Tal
Yes, but the third is only stable for set.seed(1), if you try
set.seed(250); for(i in 1:6) clustergram(swiss.x, k.range = 2:6, line.width = 0.01, add.center.points = FALSE)
then I guess you have three diagrams with one set of k=3 clusters and three with another.
But as you said, it is a difficult one so maybe that is fair and we should look at tens of diagrams. Makes me think there must be a numerical test for it that we could apply.
I like this work.
(Btw: the current version gives an error when setting the points which is why I have add.center.points = FALSE)
Hi Allan,
I fixed the problem with the center points (updated my post to it now).
Regarding the stability of the clusters – the solution you gave down is simply saying “let’s always start from the same position”.
It is good if we wanted the function to give the same results. BUT, it is hiding from us the more interesting diagnostic opportunity – which is, to see if for different seeds (cluster centers starting points), we get similar or different solutions.
So to conclude – I would rather leave this part as is 🙂
After my night sleep, I suddenly woke up with another example of this methods limitation. I’ll write about it now in the post.
Lastly – thank you for showing appreciation for this work! 🙂
Cheers,
Tal
Brilliant!!!!!!!!!!
OK, I *finally* get it! Sleeping does wonders 🙂
I recommend changing clustergram.kmeans slightly to something along the lines of:
clustergram.kmeans <-
function (Data, k, …)
{
cl <- kmeans(Data, k, nstart = 25, …)
cluster <- cl$cluster
centers <- cl$centers %*% princomp(Data)$loadings[, 1]
return(list(cluster = cluster, centers = centers))
}
This makes the diagrams essentially insensitive to the seed.
(For production code you probably want nstart to be dependent on k and NROW(Data) – I leave that as an exercise.)
Hello Allan, Thank you for the offer. Personally, I’d prefer to allow a different solution of the function for different runs for (lazy) evaluation of the stability of the clustering solution.
BTW, you could also insure to always get the same result by using
Cheers,
Tal
My biggest suggestion is that you break apart the calculation code from the plotting code. That makes the code much more reusable as others can then use the same underlying data structure but with a different visual representation.
Hello Hadley,
I took on your suggestion and made it so the “clustergram” function now relies on two other functions:
1) clustergram.kmeans – changing it will allow to implement other algorithms for clustering (such as those implemented in pam)
2) clustergram.plot.matlines – Which receives the analysis results and creates the graph. Using a different function here would allow to use it in packages such as ggplots2 (although I have no idea how to do that. But I imagine it is not difficult)
Thanks again Hadley for spending the time and attention.
With much respect,
Tal
Here’s how I’d attack the problem – http://gist.github.com/439761.
I’ve broken it down into three components:
* run the clustering algorithm and get predictions (many_kmeans and all_hclust)
* produce the data for the clustergram (clustergram)
* plot it (plot.clustergram)
I don’t think I have the logic behind the y-position adjustment quite right though.
Hello Hadley,
Thank you for the code and suggestions.
I extended this post to link to the ggplot2 implementation you wrote.
With regards,
Tal
And BTW: You should have a require(“colorspace”) in there somewhere 🙂
Hi Allan, Thanks!
I moved the COL object up, so I was left with the “require(colorspace)” somewhere in the end of the code. I fixed it (and added a warning for whoever doesn’t have the package)
Thanks for the comment on the infographic. I wish I could take credit for it. I don’t know much about clustergrams, but clustergram 6 reminds me of the brachial plexus (the plexus of nerves underneath the collarbone).
Cheers Ali 🙂
And for the others,
Martin refers to me complimenting his post here:
http://spiltmartini.com/2010/05/20/infographics/
I appreciate the initiative you took to create this pattern in R.
I need to present a paper on this subject at university.
Do you know if the generation of clustergram is already implemented in R? Where can I find the mathematical calculations that are used in Clustergram?
Thanks,
Silvana
Your presentation is very clear, even for someone with only basic knowledge of statistical programs and syntax. I have a very large data set (350,000+ observations) of national educational survey data that I need to cluster, and this implementation looks promising as a way to decide the number of clusters. It worked well for me using a small subset of the data for testing. But, how might I be able to work with missing data? When I include a larger sample data set with records with missing values, I get errors returned. Are there any solutions you could suggest? Any methods for filling in missing data that I should avoid? Thanks for any insight you can offer.
Thanks for the email. I appreciate your response. Our team has addressed how to handle the missing values and we are looking over the resulting clustergrams. We are writing up a presentation for an upcoming conference, and so please entertain another query, if you don’t mind: Please let me know, what kind of kmeans algorithm does R implement here? For example, in Matthias Schonlau’s 2004 article (Computational Statistics, 19:95-111), he displays several clustergrams using SAS, Stata, and Splus implementations of random or deterministic kmeans algorithms (pp. 102-103). Which would be most similar to what is going on here with your R code? Thanks again for your insight.
Hello Deryl,
The function presented in the post offers kmeans with it’s default configurations.
From looking at
You could find that in the details section there is the answer:
I’m liking using some alpha in the lines; I have a big dataset (300,000 points), and the alpha makes it easier to see how many lines go from one node to another.
Hi! I like this function and have two suggestions:
1. changing the noise generation command in the clustergram function to
noise <- unlist(tapply(line.width, clusters.vec, function(x){c=cumsum(x); c-mean(c)}))[order(seq_along(clusters.vec)[order(clusters.vec)])]
better arranges the lines around the center points
2. One use for color which I have found is this: If there is some grouping of the data which is known beforehand, one might want to mark the groups by these colors. (At least I did this – any one else interested?)
I am interesting to use this function for pre defined groups. Can you share it with me? Thanks
Hi Shaif,
I updated the post to make the code easier to use. Please see if that helps you.
Where is it? For you reference I have 3 groups (group1=16, g2=20 and g3=19 samples)