This is a guest post by Edwin Thoen
Currently I am doing my master thesis on multi-state models. Survival analysis was my favourite course in the masters program, partly because of the great survival package which is maintained by Terry Therneau. The only thing I am not so keen on are the default plots created by this package, by using plot.survfit. Although the plots are very easy to produce, they are not that attractive (as are most R default plots) and legends has to be added manually. I come across them all the time in the literature and wondered whether there was a better way to display survival. Since I was getting the grips of ggplot2 recently I decided to write my own function, with the same functionality as plot.survfitbut with a result that is much better looking. I stuck to the defaults of plot.survfit as much as possible, for instance by default plotting confidence intervals for single-stratum survival curves, but not for multi-stratum curves. Below you’ll find the code of the ggsurv function. Just as plot.survfit it only requires a fitted survival object to produce a default plot. We’ll use the lung data set from the survival package for illustration. First we load in the function to the console (see at the end of this post).
Once the function is loaded, we can get going, we use the lung data set from the survival package for illustration.
library(survival)
data(lung)
lung.surv <- survfit(Surv(time,status) ~ 1, data = lung)
ggsurv(lung.surv)
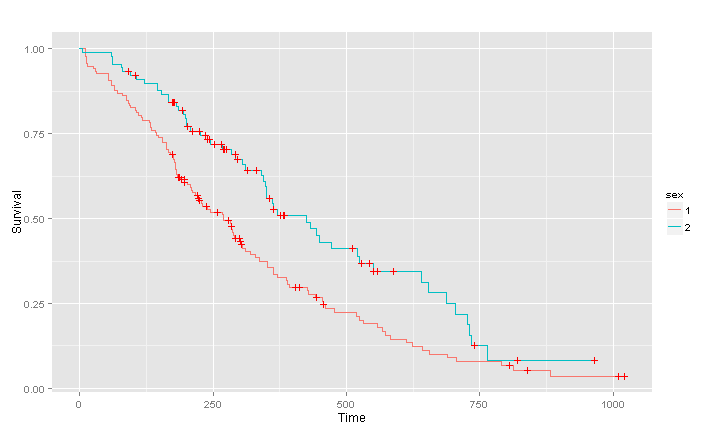
Censored observations are denoted by red crosses, by default a confidence interval is plotted and the axes are labeled. Everything can be easily adjusted by setting the function parameters. Now lets look at differences in survival between men and women, creating a multi-stratum survival curve.
lung.surv2 <- survfit(Surv(time,status) ~ sex, data = lung)
(pl2 <- ggsurv(lung.surv2))
The multi-stratum curves are by default of different colors, the standard ggplot colours. You can set them to your favourite color of course. As always with ggplots a legend is created by default. However we note that levels of the variable sex are called 1 and 2, not very informative. Fortunately the output of ggsurv can still be modified by adding layers after using the function, it is just an ordinary ggplot object.
(pl2 <- pl2 + guides(linetype = F) +
scale_colour_discrete(name = 'Sex', breaks = c(1,2), labels=c('Male', 'Female')))
That's better. Note that the function had also created a legend for linetype, that was non-informative in this case because the linetypes are the same. We removed the legend for linetype before adjusting the one for color.
Finally we can also adjust the plot itself. Maybe the oncologist is very interested in median survival of men and women. Lets help her by showing this on the plot.
lung.surv2
med.surv <- data.frame(time = c(270,270, 426,426), quant = c(.5,0,.5,0),
sex = c('M', 'M', 'F', 'F'))
pl2 + geom_line(data = med.surv, aes(time, quant, group = sex),
col = 'darkblue', linetype = 3) +
geom_point(data = med.surv, aes(time, quant, group =sex), col = 'darkblue')
I hope survival researchers will take the effort to produce better looking plots after reading this post, although copy pasting the code won't be too much of an effort I guess.
ggsurv <- function(s, CI = 'def', plot.cens = T, surv.col = 'gg.def',
cens.col = 'red', lty.est = 1, lty.ci = 2,
cens.shape = 3, back.white = F, xlab = 'Time',
ylab = 'Survival', main = ''){
library(ggplot2)
strata <- ifelse(is.null(s$strata) ==T, 1, length(s$strata))
stopifnot(length(surv.col) == 1 | length(surv.col) == strata)
stopifnot(length(lty.est) == 1 | length(lty.est) == strata)
ggsurv.s <- function(s, CI = 'def', plot.cens = T, surv.col = 'gg.def',
cens.col = 'red', lty.est = 1, lty.ci = 2,
cens.shape = 3, back.white = F, xlab = 'Time',
ylab = 'Survival', main = ''){
dat <- data.frame(time = c(0, s$time),
surv = c(1, s$surv),
up = c(1, s$upper),
low = c(1, s$lower),
cens = c(0, s$n.censor))
dat.cens <- subset(dat, cens != 0)
col <- ifelse(surv.col == 'gg.def', 'black', surv.col)
pl <- ggplot(dat, aes(x = time, y = surv)) +
xlab(xlab) + ylab(ylab) + ggtitle(main) +
geom_step(col = col, lty = lty.est)
pl <- if(CI == T | CI == 'def') {
pl + geom_step(aes(y = up), color = col, lty = lty.ci) +
geom_step(aes(y = low), color = col, lty = lty.ci)
} else (pl)
pl <- if(plot.cens == T & length(dat.cens) > 0){
pl + geom_point(data = dat.cens, aes(y = surv), shape = cens.shape,
col = cens.col)
} else if (plot.cens == T & length(dat.cens) == 0){
stop ('There are no censored observations')
} else(pl)
pl <- if(back.white == T) {pl + theme_bw()
} else (pl)
pl
}
ggsurv.m <- function(s, CI = 'def', plot.cens = T, surv.col = 'gg.def',
cens.col = 'red', lty.est = 1, lty.ci = 2,
cens.shape = 3, back.white = F, xlab = 'Time',
ylab = 'Survival', main = '') {
n <- s$strata
groups <- factor(unlist(strsplit(names
(s$strata), '='))[seq(2, 2*strata, by = 2)])
gr.name <- unlist(strsplit(names(s$strata), '='))[1]
gr.df <- vector('list', strata)
ind <- vector('list', strata)
n.ind <- c(0,n); n.ind <- cumsum(n.ind)
for(i in 1:strata) ind[[i]] <- (n.ind[i]+1):n.ind[i+1]
for(i in 1:strata){
gr.df[[i]] <- data.frame(
time = c(0, s$time[ ind[[i]] ]),
surv = c(1, s$surv[ ind[[i]] ]),
up = c(1, s$upper[ ind[[i]] ]),
low = c(1, s$lower[ ind[[i]] ]),
cens = c(0, s$n.censor[ ind[[i]] ]),
group = rep(groups[i], n[i] + 1))
}
dat <- do.call(rbind, gr.df)
dat.cens <- subset(dat, cens != 0)
pl <- ggplot(dat, aes(x = time, y = surv, group = group)) +
xlab(xlab) + ylab(ylab) + ggtitle(main) +
geom_step(aes(col = group, lty = group))
col <- if(length(surv.col == 1)){
scale_colour_manual(name = gr.name, values = rep(surv.col, strata))
} else{
scale_colour_manual(name = gr.name, values = surv.col)
}
pl <- if(surv.col[1] != 'gg.def'){
pl + col
} else {pl + scale_colour_discrete(name = gr.name)}
line <- if(length(lty.est) == 1){
scale_linetype_manual(name = gr.name, values = rep(lty.est, strata))
} else {scale_linetype_manual(name = gr.name, values = lty.est)}
pl <- pl + line
pl <- if(CI == T) {
if(length(surv.col) > 1 && length(lty.est) > 1){
stop('Either surv.col or lty.est should be of length 1 in order
to plot 95% CI with multiple strata')
}else if((length(surv.col) > 1 | surv.col == 'gg.def')[1]){
pl + geom_step(aes(y = up, color = group), lty = lty.ci) +
geom_step(aes(y = low, color = group), lty = lty.ci)
} else{pl + geom_step(aes(y = up, lty = group), col = surv.col) +
geom_step(aes(y = low,lty = group), col = surv.col)}
} else {pl}
pl <- if(plot.cens == T & length(dat.cens) > 0){
pl + geom_point(data = dat.cens, aes(y = surv), shape = cens.shape,
col = cens.col)
} else if (plot.cens == T & length(dat.cens) == 0){
stop ('There are no censored observations')
} else(pl)
pl <- if(back.white == T) {pl + theme_bw()
} else (pl)
pl
}
pl <- if(strata == 1) {ggsurv.s(s, CI , plot.cens, surv.col ,
cens.col, lty.est, lty.ci,
cens.shape, back.white, xlab,
ylab, main)
} else {ggsurv.m(s, CI, plot.cens, surv.col ,
cens.col, lty.est, lty.ci,
cens.shape, back.white, xlab,
ylab, main)}
pl
}




Great post. Is it also possible to use ggsurf if I use glm or glmer to estimate a (multi-level) discrete-time hazard model (see http://www.ats.ucla.edu/stat/r/examples/alda/ch12.htm)?
The function only works if it is used on an object of class survfit. If you want it to work on a different object you should tweak the code a bit. Note that the first part of the function is creating data frames that are fed to the ggplot code below. If you can turn your fit into a data frame just alike you can readily use the code that produces the plots. Good luck!
See also slide 68 onwards in http://timchurches.github.io/ggplot2er/ which illustrates the use of a similar approach by Ramon Saccilotto of the Basel Institute for Clinical Epidemiology and Biostatistics (links to Ramon’s work are in the presentation).
Didn’t see that one before, thanks. Decided to write the function because I couldn’t find any function or code. Nice slides by the way, Lung data set is popular!
Either your function or Ramon’s (no idea which one works best) should get submitted to the GGally package, or perhaps even to the autoplot package.
Thanks for the suggestion, I will look into the options.
No worries. I’ll suggest little things if you submit it to GGally, like vectorizing the strata loops or leaving out the xlab and ylab arguments to encourage the use of the labs() function in ggplot2.
Back with another suggestion: Christopher Gandrud’s simPH package seems very relevant here. It plots everything with ggplot2 and has a ggfitStrata function.
See also slide 68 onwards in http://timchurches.github.io/ggplot2er/ which illustrates the use of a similar approach by Ramon Saccilotto of the Basel Institute for Clinical Epidemiology and Biostatistics (links to Ramon’s work are in the presentation).
While these are a nice example of using ggplot syntax, you might want to also look at the features associated with survplot() in the rms package by Frank Harrell – all based in base graphics. The confidence bands are worth their weight in gold.
The survival rate in my data set is 20%. Following your syntax, the range of the y-axis was constrained between 0.75 and 1.00. So what if I want the y-axis ranges from 0 to 1?Thank you!
You can just use the ggplot2 function ylim to adjust the y-axis
ggsurv(my.survfit) + ylim(0, 1)
Although non-R, there is also a win-based tool to draw survival time plots:
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0038960
Holger
Hoping to solicit some help re: two issues with ggsurv. My plot contains two strata similar to the second example where you stratify by sex.
1: how can you redefine the linetype of the KM curves using ggsurv? I want to make them dashed (for example)/ Using linetype to modify these curves does not seem to work.
2: I can overlay a smoothed line on top of the KM step-plot by adding: + geom_line() to the code. However, the colors do not correspond to the colors of the step curve. How do I force ggplot to make them the same color as their respective step curve?
Thanks for your help,
Bob
Hi Bob,
Thanks for your questions.
1) There is a built in option for the line type of the estimates
ggsurv(s, lty.est = 2)
would give a dashed line for the survival curves.
2) Within the ggsurv function the different strata levels are stored in the variable called group. This variable is used to create the different colors for the strata. You can use this variable also if you want to make additions to the plot. (I would suggest to use the smooth geom instead of the line geom, if you want to add a smoother to the plot)
survPlot <- ggsurv(s)
survPlot + geom_line(aes(color = group))
survPlot + geom_smooth(aes(color = group), se = F)
Good luck,
Edwin
Can you help me with adjusting the size of curves?
Also, how can I set the colors of multi-stratum curves?
Thank you for your help.
Jin.
Hi Jin,
You can specify the colors at the surv.col argument. Just enter a vector with color names of the same length as the number of strata, for example c(“red”, “green”). There is no argument to specify line width, but you are free to add that to the function in the code above of course!
Good luck,
Edwin
Thank you very much for your help.
Could you write the added code. I cannot figure it out to add width.
Thanks for sharing this general solution for plotting survival curves with multiple strata. I just want to suggest a couple things about the code. It seems cleaner to separate the helper functions ggsurv.s and ggsurv.m instead of redefining them every time ggsurv is called; this can be done cleanly without relying on the currently shared variable “strata” in the parent function (e.g., just define it again in ggsurv.m where it’s used). It also seems cleaner to use a consistent type for function arguments (e.g., “CI” could be logical or character) to simplify checks (e.g., “if(CI)”). It’s completely unnecessary to compare logical variables with truth (e.g., “if(CI)” is simpler and clearer than “if(CI == TRUE)”). It seems you are mixing curly braces and parentheses in your if-then statements. I think it’s easier to read assignments inside if-then statements instead of assigning the result of the entire if-then statement to an object. It’s simpler and less error-prone to rely on data.frame in-built mechanism to repeat an atomic value the appropriate number of times (e.g., “data.frame(x=1:10, group=groups[i])” instead of “data.frame(x=1:10, group=rep(groups[i], n[i]+1))”). Using “with” avoids repeatedly accessing various slots from within the same object (e.g., “s$time”, “s$surv”, “s$upper”, etc.). It seems preferable to write out “TRUE” and “FALSE” instead of relying on “T” and “F” which can be overridden in some environments. And I think you mean “black.white” instead of “back.white”. Again, thanks for sharing your code. I intend to use it following a bit of clean up.
Hello! Can you share your cleaned up code to us? Thank you!
Thanks for your suggestions Roman, I certainly don’t regard myself as a professional R programmer, so all suggested improvements to my coding are more than welcome! Currently I am working on getting the function to the GGally package and adjustments to the original code already have been made. I will keep your comments in mind when further improving the code. Thanks again!
How do I make the axes’ values more discrete?
I have my y-axis only display 1.0, 0.9, 0.8 and I would like it to be more specific ie. 1.0, 0.95, 0.9, 0.85, 0.8 at least.
I am sorry I didn’t notice your comment earlier. The plots produced by ggsurv() are just ggplot opbjects, so you can apply the ggplot function scale_y_continous() in this case. Assuming that you have saved your plot in an object called p:
p + scale_y_continuous(breaks = seq(0, 1, by = .1))
Good luck,
Edwin
Is there a way to add shading to each of the strata’s confidence intervals. Perhaps, taking it a bit farther, applying a different shade color to the regions of overlap, i.e one strata will be blue and another red then the overlap region could be purple? Just some thoughts
Thanks for the suggestion, I would imagine we would use the “ribbon” geom for this. Unfortunately I have no time to look into it right now but you are absolutely welcome to tweak the code to do this.
hi, quick question, why you have red dots on your male line (which is green)?
The default color for the censored observations is red, irrespective of the color of the line. This can be adjusted by entering the color of your liking at cens.col.
Hi, this is really impressive work. Just wanna ask is it possible to adjust the “censored” dots to the same color as its own curve? That is, make the “male” dots green while the “female” dots keep red at the same time. Thanks.
Thank you for your question KW87. Answer is unfortunately “no, it is not”. Since more people did ask for it I will accommodate for it in upcoming version of ggsurv. Whenever GGally updates. Remind me if you don’t see this option appear in a while.
Cheers,
Edwin
I am curious if you have gotten around to allowing different colors for censor marks. I really appreciate all the work you have put into ggsurv!
I am happy to inform you all that the ggsurv function is now available in the GGally package.
install GGally but ggsurv is still not available any reason?
Thanks for notifying. I just contacted the package admin Barret, something went wrong with the publication of the latest package version. It will be back in at the next version, which will be released soon. Sorry for this…
Any idea just about when soon is? I like the concept of homogenizing all my plots in a paper, to a “ggplot” format…so the survival plot with your function would be great to have! Thanks for the code by the way!
I am not sure, Barret is on the road right now. He would update the package as soon as he would get back. I’ll expect it any day now. In the mean time you can still copy paste the above code to produce your plots for the paper. Good luck!
Cheers. I’ll give it a shot. Is this code identical to whats included in the GGally package? Thanks also for your fast reply. Oliver
The code in the package is adjusted so it is in the same style as the rest of the package. Function output of the above code should be exactly identical to the output of the code however.
So I ran the code on a survival function, I’d made by stratifying groups by expression data in my data.frame.
survexp =2200
survexpdif 1 and only the first element will be used
Hide Traceback
Rerun with Debug
Error in unlist(strsplit(names(s$strata), “=”)) :
error in evaluating the argument ‘x’ in selecting a method for function ‘unlist’: Error in strsplit(names(s$strata), “=”) : non-character argument
4 unlist(strsplit(names(s$strata), “=”))
3 factor(unlist(strsplit(names(s$strata), “=”))[seq(2, 2 * strata,
by = 2)])
2 ggsurv.m(s, CI, plot.cens, surv.col, cens.col, lty.est, lty.ci,
cens.shape, back.white, xlab, ylab, main)
1 ggsurv(survexpdif)
Any idea what I did wrong?
Cheers,
O
Mmm, this should work. Just to make sure, is the survival object correct? Summary stats and ordinary KM plot do work? Otherwise it is difficult to say at distance, maybe you can share your data somehow (or make up data in the same form) so I can have a look.
Yeah, the survival object works normally in R. Normal summary stats, can plot it also using the plot() function. Undersatnd its diff to run without a reproducible example. Will have to think how I can generate a similar dataset. Suck at making data in R :-). I can otherwise email you the txt file. But thanks for your help until now.
Could you please send the data and a script in which you prep the dat upto the point you just describe to
edwinthoen at gmail dot com.
Thanks,
Edwin
isn’t it possible to use it for the coxph models? if not, do you have suggetions to where I can find nice graph codes for this?
what i mean is: i want to plot survival curves for specific values of my two covariates e.g. covariate 1=1(binary) and covariate 2=56716(continuous). I have tried to use your code, but it doesn’t work. I’ve plottet it like this until now, but it doesn’t look pretty:
fitCox <- coxph(Surv(days, ms) ~ chi + strata(MR), retro85tilr)
tempNewData <- data.frame(chi=c(56716), MR=c(1))
plot(survfit(fitCox, newdata=tempNewData),conf.int=T)
can you help?
Sorry for the late reply. Unfortunately the dataset you use is not publicly available (I wasn’t able to find it at least). However reproducing on the lung data set, I think this is what you are after. Please let me know if it is not what you are looking for.
fitCox <- coxph(Surv(time, status) ~ age + strata(sex), lung)
tempNewData <- data.frame(age=c(60), sex=c(1))
fitSurv <- survfit(fitCox, newdata=tempNewData)
ggsurv(fitSurv, CI = TRUE)
First, thank you for this! I *definitely* prefer these plots!
However, I followed your instructions above for plotting a Cox PH model using new data (using my own data set), and I keep getting the error below. What am I doing wrong?
Error in grid.Call.graphics(L_lines, x$x, x$y, index, x$arrow) :
invalid line type
Hi Matt
I have the same problem. Have you solved this problem. Can you post you code so we can debug what is going on?
Hi Edwin
Have you tested this code? This code will not work for sure. I will get the error mentioned by Matt
Hi,
I am looking to investigate the interplay of Overall and Progression free survival with respect to treatment and control arm to estimate the relationship between survival curve and cost effectiveness. I do not have patient level data but wanted to do theoretical modelling using 6 different shapes of OS and PFS but choice of which distribution to use (Justification) seems to be a problem!!! Can you help in this regard?
Syed
[email protected]
That sounds like a theoretical problem, nog a plotting issue. I cannot give you a straight answer to that, if you are looking for distributions for survival curves you best consult a textbook on survival analysis like the one Klein & Moeschberger.
Good luck,
Edwin
Hi,
Could you show me an example how to plot more than 2 survival curves using your function please?
Thank you
Hey
Thanks a lot! Is it also possible to inverse the plot?
Thanks in advance
Nina
Dear Edwin, I’m making an “inverse” survival curve using your code. I realized that in order to make such graph, I have to edit the y axis information in your ggplot comments from “y=surv” into “y=1-surv”. So if I already installed GGally package, is there any support or suggestion on how to make inverse graph without the need of editing the code?
Thank you so much
Nguyet
It’s not perfect, but with a little bit of hacking in the survival object you come a long way. Maybe I’ll integrate into upcoming versions. Good luck! Edwin
library(GGally)
library(magrittr)
survival.inverse <- function(sf.object){
sf.return <- sf.object
sf.return$surv <- 1 – sf.return$surv
sf.return$upper <- 1 – sf.return$upper
sf.return$lower <- 1 – sf.return$lower
sf.return
}
scrape.off.zero % max %>% add(1))
}
sf.lung <- survival::survfit(Surv(time, status) ~ 1, data = lung)
sf.lung.inv <- survival.inverse(sf.lung)
plot1 % scrape.off.zero
sf.sex <- survival::survfit(Surv(time, status) ~ sex, data = lung)
sf.sex.inv <- survival.inverse(sf.sex)
plot2 % scrape.off.zero
I thought you might want to try cens.shape=124, the ‘|’ character instead of “+”. It looks a bit more like the actual censor mark. I’m still searching for a good way to put an actual censor mark (vertical, above the survival line).
Hi, I have a data set that is looking at, not just sex, but species (of dragonfly) and size of prey attacked. My model is:
dfsurvival3<- survfit(Surv(time, event) ~ group + genderf + beadf)
Gender and bead are as.factor, group is species. How would I edit the code to show all 8 lines? I'm sorry if this is a silly question, I'm fairly new to R and coding, in general. If you could help, that would be greatly appreciated.
What do You mean by 8 lines? Do You mean that You have 2 groups, 2 beads and 2 genders and want to draw all possible combinations?
Thank you, thank you, thank you!!!
Does anybody know how to assign different textures to line within one graph for the survfit function?
If You use the modern version of ggsurv from the package ggally you can specify the texture of the line directly in the ggsurv call with lty.est
For example
p1 <- ggsurv(fit, plot.cens=FALSE, lty.est=c(1,2,4), surv.col=c("black","dark blue", "red" ))
Actually, the ggsurv in GGallypackage doesn’t provide the choice for changing line width as well. The parameter you used is just for altering the line type.
I’ve pulled a request on ggally, hoping they could add it asap.
When I work my way through this tutorial, I get hung up on the part where you add legends. For some reason, I get the error message:
“could not find function ‘scale_colour_discreet’ ”
…even though R-Studio suggests this function as an auto-complete option. I guess this is some kind of dependencies issue; I’m working with the following packages installed:
– survival
– ggplot2
– GGally
Any ideas as to why I’m getting this error message?
How can you plot Hazard function using ggsurv? Or more generally, how can you plot a curve after some transformation of the survival curve. For example, in the original ‘plot’ function, you can do plot(survfitModel$time, log(-log(survfitModel$survival))), ‘survfitModel’ is my survfit object. Can you do similar thing in ggsurv?
This my ‘survDataFrame’ data set
DeviceId Status SurvivalTime DeviceAgeWhenEnterStudy MDC1DeviceFamily MDC2FormFactor
1 g:6755399441324328 1 46 498 PC Desktop
2 g:6755399445788225 1 42 491 PC Notebook
3 g:6755399446628345 0 334 620 PC Notebook
4 g:6755399446805200 0 334 254 PC Notebook
IsTouchEnabled CustomerGroup VMConfiguration IsOEMNewWin10 Region
1 True Commercial – Other Host only Upgraded United States
2 False Consumer Host only Upgraded Greater China
3 False Gaming Host only Upgraded Western Europe
4 False Gaming Host only Upgraded Latam
I first fit the coxph model and then plot the figure using ‘ggsurv’
survFitCoxPHByUpgrade = coxph(survObj ~ MDC1DeviceFamily + strata(IsOEMNewWin10) , data = survDataFrame);
tempData = data.frame(MDC1DeviceFamily = “PC”) ;
survFitByUpgrade = survfit(survFitCoxPHByUpgrade, newdata = tempData);
ggsurv(survFitByUpgrade, lty.est = 1, lty.ci = 2, size.est = 2, size.ci = 0.5) + theme(text = element_text(size = 20)) + theme(axis.text = element_text(size = 10));
I got the following error
“Error in grid.Call.graphics(L_lines, x$x, x$y, index, x$arrow) :
invalid line type”
However, it works fine if I use plot((survFitByUpgrade).
What is going on?
I am throwing the same error when I attempt to plot my survival object: “Error in grid.Call.graphics(L_lines, x$x, x$y, index, x$arrow) :
invalid line type”
I have searched the function and there are numerous calls to lty, and I am not sure which is causing this error. Any suggestions on moving forward? I really like the looks of your stratified plots and am hoping to produce something similar.
For those who are still looking here for nice survival plots, please look at https://www.r-bloggers.com/survival-plots-have-never-been-so-informative/
HTH
Hi Edwin, Many thanks for this code. Can you please tell me how to compute and plot the log rank value on the lower left side of the plot?
THanks