This post shows how to print a prettier nested pivot table, created using the {reshape} package (similar to what you would get with Microsoft Excel), so you could print it either in the R terminal or as a LaTeX table. This task is done by bridging between the cast_df object produced by the {reshape} package, and the tabular function introduced by the new {tables} package.
Here is an example of the type of output we wish to produce in the R terminal:
1 2 3 4 5 6 7 | ozone solar.r wind temp month mean sd mean sd mean sd mean sd 5 23.62 22.22 181.3 115.08 11.623 3.531 65.55 6.855 6 29.44 18.21 190.2 92.88 10.267 3.769 79.10 6.599 7 59.12 31.64 216.5 80.57 8.942 3.036 83.90 4.316 8 59.96 39.68 171.9 76.83 8.794 3.226 83.97 6.585 9 31.45 24.14 167.4 79.12 10.180 3.461 76.90 8.356 |
Or in a latex document:
Motivation: creating pretty nested tables
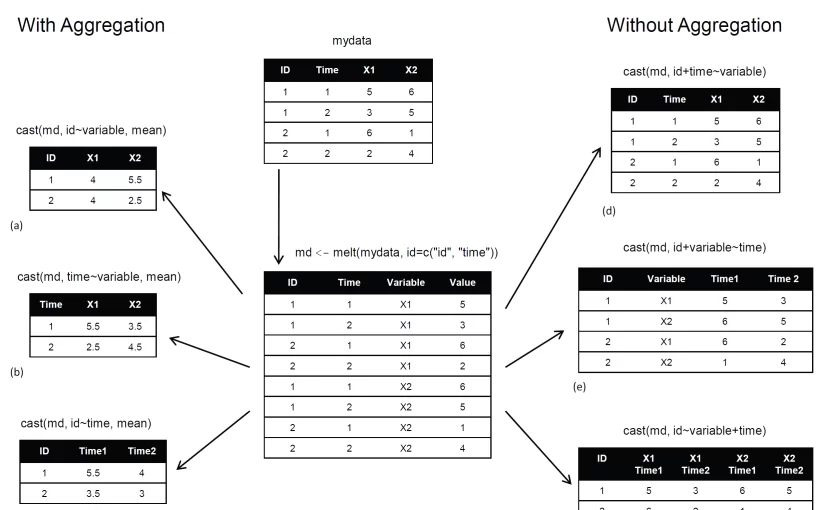
In a recent post we learned how to use the {reshape} package (by Hadley Wickham) in order to aggregate and reshape data (in R) using the melt and cast functions.
The cast function is wonderful but it has one problem – the format of the output. As opposed to a pivot table in (for example) MS excel, the output of a nested table created by cast is very “flat”. That is, there is only one row for the header, and only one column for the row names. So for both the R terminal, or an Sweave document, when we deal with a more complex reshaping/aggregating, the result is not something you would be proud to send to a journal.
The opportunity: the {tables} package
The good news is that Duncan Murdoch have recently released a new package to CRAN called {tables}. The {tables} package can compute and display complex tables of summary statistics and turn them into nice looking tables in Sweave (LaTeX) documents. For using the full power of this package, you are invited to read through its detailed (and well written) 23 pages Vignette. However, some of us might have preferred to keep using the syntax of the {reshape} package, while also benefiting from the great formatting that is offered by the new {tables} package. For this purpose, I devised a function that bridges between cast_df (from {reshape}) and the tabular function (from {tables}).
The bridge: between the {tables} and the {reshape} packages
The code for the function is available on my github (link: tabular.cast_df.r on github) and it seems to works fine as far as I can see (though I wouldn’t run it on larger data files since it relies on melting a cast_df object.)
Here is an example for how to load and use the function:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 | ###################### # Loading the functions ###################### # Making sure we can source code from github source("https://www.r-statistics.com/wp-content/uploads/2012/01/source_https.r.txt") # Reading in the function for using tabular on a cast_df object: source_https("https://raw.github.com/talgalili/R-code-snippets/master/tabular.cast_df.r") ###################### # example: ###################### ############ # Loading and preparing some data require(reshape) names(airquality) <- tolower(names(airquality)) airquality2 <- airquality airquality2$temp2 <- ifelse(airquality2$temp > median(airquality2$temp), "hot", "cold") aqm <- melt(airquality2, id=c("month", "day","temp2"), na.rm=TRUE) colnames(aqm)[4] <- "variable2" # because otherwise the function is having problem when relying on the melt function of the cast object head(aqm,3) # month day temp2 variable2 value #1 5 1 cold ozone 41 #2 5 2 cold ozone 36 #3 5 3 cold ozone 12 ############ # Running the example: tabular.cast_df(cast(aqm, month ~ variable2, c(mean,sd))) tabular(cast(aqm, month ~ variable2, c(mean,sd))) # notice how we turned tabular to be an S3 method that can deal with a cast_df object Hmisc::latex(tabular(cast(aqm, month ~ variable2, c(mean,sd)))) # this is what we would have used for an Sweave document |
And here are the results in the terminal:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | > > tabular.cast_df(cast(aqm, month ~ variable2, c(mean,sd))) ozone solar.r wind temp month mean sd mean sd mean sd mean sd 5 23.62 22.22 181.3 115.08 11.623 3.531 65.55 6.855 6 29.44 18.21 190.2 92.88 10.267 3.769 79.10 6.599 7 59.12 31.64 216.5 80.57 8.942 3.036 83.90 4.316 8 59.96 39.68 171.9 76.83 8.794 3.226 83.97 6.585 9 31.45 24.14 167.4 79.12 10.180 3.461 76.90 8.356 > tabular(cast(aqm, month ~ variable2, c(mean,sd))) # notice how we turned tabular to be an S3 method that can deal with a cast_df object ozone solar.r wind temp month mean sd mean sd mean sd mean sd 5 23.62 22.22 181.3 115.08 11.623 3.531 65.55 6.855 6 29.44 18.21 190.2 92.88 10.267 3.769 79.10 6.599 7 59.12 31.64 216.5 80.57 8.942 3.036 83.90 4.316 8 59.96 39.68 171.9 76.83 8.794 3.226 83.97 6.585 9 31.45 24.14 167.4 79.12 10.180 3.461 76.90 8.356 |
And in an Sweave document:
Here is an example for the Rnw file that produces the above table:
cast_df to tabular.Rnw
I will finish with saying that the tabular function offers more flexibility then the one offered by the function I provided. If you find any bugs or have suggestions of improvement, you are invited to leave a comment here or inside the code on github.
(Link-tip goes to Tony Breyal for putting together a solution for sourcing r code from github.)