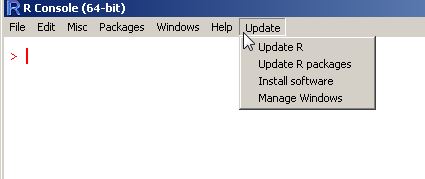
Upgrading R on Windows is not easy. While the R FAQ offer guidelines, some users may prefer to simply run a command in order to upgrade their R to the latest version. That is what the new {installr} package is all about.
The {installr} package offers a set of R functions for the installation and updating of software (currently, only on Windows OS), with a special focus on R itself. To update R, you can simply run the following code:
# installing/loading the package:
if(!require(installr)) {
install.packages("installr"); require(installr)} #load / install+load installr
# using the package:
updateR() # this will start the updating process of your R installation. It will check for newer versions, and if one is available, will guide you through the decisions you'd need to make.
Running this function will perform the following steps:
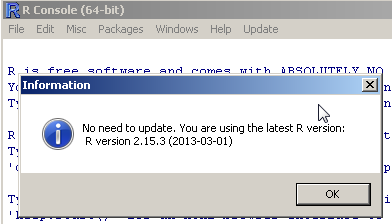
Check what is the latest R version. If the current installed R version is up-to-date, the function ends (and returns FALSE)
If a newer version of R is available, you will be asked if to review the NEWS of the latest R version – in order to decide if to install the
newest R or not.
If you wish it – the function will download and install the latest R version. (you will need to press the "next" buttons on your own)
Once the installation is done, you should press "any-key", and the function will proceed with copying all of your packages from your old (well, current) R installation, into your newer R installation.
You can then erase all of the packages in your old R installation.
After your packages are moved (and the old ones possibly erased), you will get the option to update all of your packages in the new version of R.
Lastely – you can open the new Rgui and close the current session of your old R. (This is a bit buggy in version 0.8, but has been fixed in version 0.8.1)
If you know you wish to upgrade R, and you want the packages moved (not copied, MOVED), you can simply run:
# installing/loading the package:
if(!require(installr)) { install.packages("installr"); require(installr)} #load / install+load installr
updateR(F, T, T, F, T, F, T) # install, move, update.package, quit R.
Since the various steps are broken into individual functions, you can also pick and choose what to run using the relevant function:
# installing/loading the package:
if(!require(installr)) { install.packages("installr"); require(installr)} #load / install+load installr
# step by step functions:
check.for.updates.R() # tells you if there is a new version of R or not.
install.R() # download and run the latest R installer
copy.packages.between.libraries() # copy your packages to the newest R installation from the one version before it (if ask=T, it will ask you between which two versions to perform the copying)
If you like using the global library system, you can run the following in the old R:
# installing/loading the package:
if(!require(installr)) { install.packages("installr"); require(installr)} #load / install+load installr
updateR(F, T, F, F, F, F, T) # only install R (if there is a newer version), and quits it.
And then run the following in the new version of R:
source("https://www.r-statistics.com/wp-content/uploads/2010/04/upgrading-R-on-windows.r.txt")
New.R.RunMe()
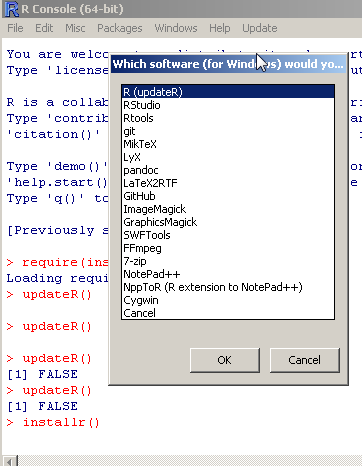
The {installr} package also offers functions for installing various other software on Windows. These functions include: install.pandoc (which was mentioned on this blog recently), install.git, install.Rtools, install.MikTeX, install.RStudio, and a general install.URL and install.packages.zip functions. You can see these further explained in the package’s Reference manual.
Feature requests, bug reports – and your help in improving the package
You can see the latest version of installr on github, where you can also submit bug reports (you may also just leave a comment in this post). Since this is my first R package, I might have (e.g: probably have) missed something here or there. So any comment on how to improve my code/documentation/R-fu, will be most welcomed (here or on github).
If this type of coding is fun/easy for you, you can help me improve this package on github. Cool new features I think may be added (by me or others) are:
- Add an uninstall.R function – to remove the old R version.
- Add more support for upgrading R for people who uses a global library for their packages.
- Add support for Linux and Mac! This one I am less likely to do on my own – and would love to see someone else extend my code to other operation systems.
- GUI – add a menu based option for running updateR. Something like help->”check for updates” would be great. (p.s: this idea came from Yihui Xie)
- add even more install.software functions. If you have functions for which you’d like to be able to easily install them – just let me know and it could be included in future releases.
Thanks
Final note, I would like to thank the many people who have developed WONDERFUL tools for making R package development possible (and even somewhat fast), on Windows. These include Prof. Brian Ripley and Duncan Murdoch for Rtools, also Uwe Ligges for his work on CRAN, Hadley Wickham for devtools (in general, and for its documentation), Yihui Xie for roxygen2, JJ and others in the RStudio team for RStudio, the people behind git and github, and more. There are probably more things I can thank these people for, and many more people I should thank, but I can’t figure who you are probably (feel free to e-mail me, I appreciate you work even if it is not clear to me your are behind it).